
Source: Sonia Negrao, UCD
Barley is one of the main crops in Europe regarding cultivated area, economic output and agroecological impact. Barley success relies on an extraordinary genetic diversity that allowed its adaptation to environments far removed geographically and climatically from its centre of origin. However, future breeding must respond to the sum of challenges posed by a changing climate, increasing resource limitation, and the societal mandate for sustainable production and environment-aware agriculture. Plant scientists must rethink the processes that shaped crop adaptation, harness the wealth of scientific knowledge generated during the genomic revolution, and re-start a data-driven exploitation of genetic diversity to harness new diversity based on knowledge acquired.
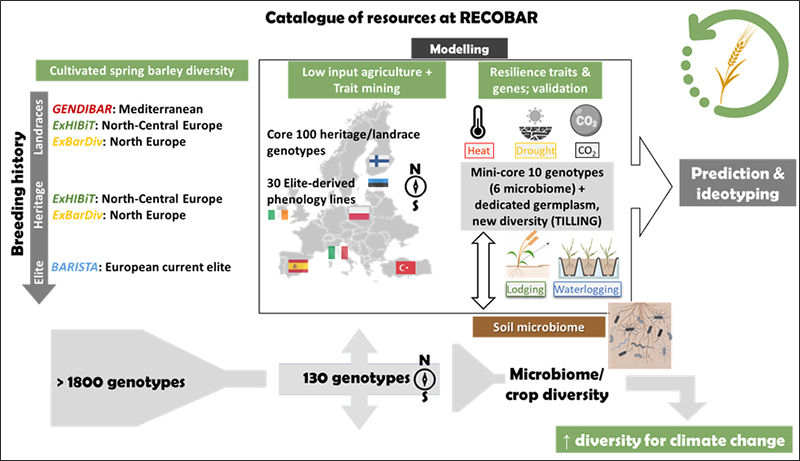
This project builds on a rich catalogue of germplasm, a variety of approaches addressing the use of barley diversity in novel ways, and a solid foundation of previous knowledge (genotypic, phenotypic and functional) built during past collaborations.
We propose a variety of approaches to broaden barley's genetic base and its impact at different scales: exploration of the wide diversity harboured by old varieties and landraces for adaptation to shifting climates, discovery and deployment of genes affecting key traits, exploration of targeted modification of known genes, improvement of functional crop growth models applied to genomic prediction with newly unravelled genetic diversity, and evaluation of soil microbiota diversity dynamics in relation to barley diversity.
Source: Sonia Negrao, UCD
The project addresses the following challenges:
Harnessing extant barley diversity to expand the crop genetic base. Genetic diversity lost due to modern breeding will be explored. The potential for direct use of landraces from the Mediterranean basin, Nordic-Baltic region, and European legacy cultivars in low input agriculture will be studied by field testing of mini-core collections. Best accessions will be promoted for pre-breeding. Suitability of barley lines with modified phenology for shifting agroecosystems. Double cropping is already practised in Europe, but can be further optimised with the development of short season winter cereal crops, opening more options for crop rotations, promoting more resilient agroecosystems. The project will study a selection of already developed introgression lines after a worldwide exploration of germplasm, enriching the phenology responses of elite germplasm. These activities will be disseminated to stakeholders like farmer associations and breeding companies.

Source: : Ernesto Igartua Arregui, CSIC
Mining new traits and QTL for tolerance to abiotic stresses with new and old diversity. The project will work with a variety of plant materials already developed by consortium partners to improve barley for responses to drought, waterlogging, high temperature, low N, lodging. Landrace and legacy germplasm will be tested for high temperature and waterlogging stresses, using state-of-the-art phenotyping method, and root phenotyping of outstanding genotypes.
Barley ideotyping for future agroecosystems. The most cost-effective way to test plant innovations are model-aided approaches. Models must become truly functional to become predictive tools. New models for simulation of biotic and abiotic stress impacts in barley growth will be developed. Models will be used to extend GP under climate change scenarios and genotypic responses to shifting agroecosystems, and to identify - using data from the germplasm evaluation tasks - region-specific breeding toolkits.
Rhizosphere microbiota associations with crop diversity and environmental conditions. Microbiota composition responds to environmental conditions, crop management and specific cultivar. Plant domestication and modern breeding have impacted on the composition and functions of the rhizosphere microbiota. Soil microbiomics, and AMF colonisation will be explored in field trials of selected cultivars with different breeding histories, under a variety of environments.

Source: : Ernesto Igartua Arregui, CSIC
Ernesto Igartua Arregui
CSIC, Aula Dei Experimental Station, Spain
Email: igartua@eead.csic.es
Alan Schulman
University of Helsinki, Finland
Hakan Ozkan
University of Cukurova, Türkiye
Laura Rossini
University of Milano, Italy
Alessandro Tondelli
CREA, Research Centre for Genomics and Bioinformatics, Italy
Agata Daszkowska-Golec
University of Silesia in Katowice, Poland
Hannes Kollist
University of Tartu, Estonia
Sirja Viitala
Natural Resources Institute, LUKE, Finland
Sonia Negrao
University College Dublin, Ireland