Advanced tools for breeding BARley for Intensive and SusTainable Agriculture under climate change scenarios
Source: Luigi Cattivelli
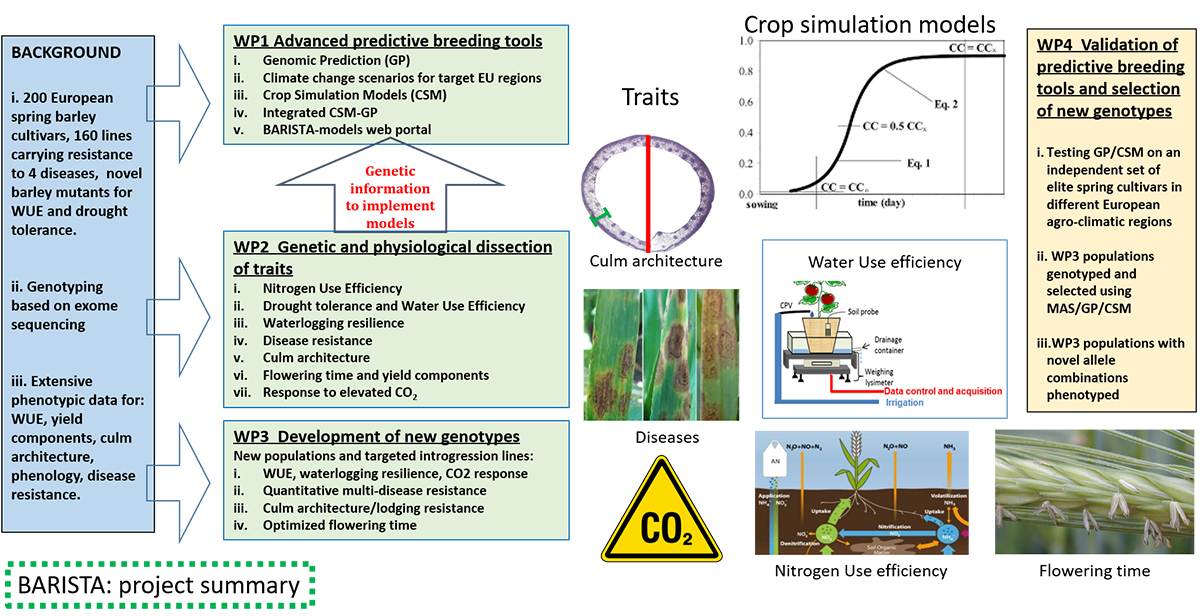
Improving the genetic potential of the seeds is the most effective way to introduce the innovation in agriculture needed to meet the UN Sustainable Development Goals (SDGs). Plant breeding allows improvement of crop sustainability and yield potential through the introgression of specific traits capable of coping with climate change, disease shifts, and resource limitations. BARISTA is delivering new breeding strategies and toolkits for boosting barley crop improvement, leading to new, high-yielding varieties selected to cope with anticipated future climatic conditions.
BARISTA is built on extensive phenotypic and genotypic data generated in previous projects and on current understanding of the genetics of ideotype traits for biotic and abiotic stress resilience. The consortium works on a common set of germplasm, consisting of ~200 barley spring cultivars extensively genotyped and phenotyped within the previous Exbardiv (ERA-PG 2006), Climbar (FACCE-JPI 2014), and various national projects, a panel of 160 spring barley single- and quadruple-stack lines developed to introgress quantitative resistance against the main barley pathogens, and several novel barley ABA-related mutants of candidate genes affecting water use efficiency (WUE) and drought tolerance.

Source: Luigi Cattivelli
In BARISTA, this germplasm and data resource is driving genomic prediction (GP) and crop simulation models (CSMs) in combination to improve current predictive breeding tools and methods. In addition, we are dissecting traits relevant for barley sustainability and resilience (e.g., water- and nitrogen-use efficiency, culm architecture, disease resistance, flowering time) by using state of the art phenotyping, genetics, and genomics methodologies.
BARISTA is working to release a barley breeding toolkit made of crop simulation models capable to predict crop performance under different climatic scenario and novel genes/alleles for sustainable production. The following major research findings and outputs have been achieved so far:
In terms of crop simulation models:
In terms of new genes and genotypes the project has been identified or selected:
Overall, BARISTA provides tools for breeding in general and genes/genotypes for improving crop sustainability (e.g. diseases resistance, nitrogen use efficiency, water use efficiency).

Source: Luigi Cattivelli
Dr Luigi Cattivelli
Consiglio per la ricerca in agricoltura e l'analisi dell'economia agraria (CREA)-Research centre for genomics and bioinformatics, ITALY
Email: luigi.cattivelli@crea.gov.it
Prof Laura Rossini
Università degli Studi di Milano, Department of Agricultural and Environmental Sciences - Production, Landscape, Agroenergy (DiSAA), ITALY
Prof Alan Schulman
Luonnonvarakeskus (Luke), Production Systems, FINLAND
Dr Ana M Casas
Estación Experimental Aula Dei-CSIC, Genetics and Plant Production, SPAIN
Prof Klaus Pillen
Martin-Luther-University Halle/Wittenberg, Institute of Agricultural and Nutrional Sciences, GERMANY
Prof Reimund P Roetter
University of Goettingen, Department of Crop Sciences, GERMANY
Prof Soren K. Rasmussen
University of Copenhagen, Department of Plant and Environmental Sciences, DENMARK
Prof Robbie Waugh
James Hutton Institute, UK
Prof Hannes Kollist
University of Tartu, Institute of Technology, ESTONIA
Dr Agata Daszkowska-Golec
University of Silesia in Katowice, Faculty of Biology and Environmental Protection, Department of Genetics, POLAND
Dr Leif Knudsen
SEGES Landbrug & Fødevarer F.m.b.A. Planteinnovation, DENMARK
Dr Mati Koppel
Estonian Crop Research Institute, ESTONIA
2023
Appiah, M., Bracho-Mujica, G., C.R. Ferreira, N., H. Schulman,A., P. Rötter, R. (2023). Projected impacts of sowing date and cultivar choice on the timing of heat and drought stress in spring barley grown along a European transect. Field Crops Research 291, 108768.
Paul, M., Tanskanen, J., Jääskeläinen, M., Chang, W., Dalal, A., Moshelion, M., and Schulman, A. H. (2023). Drought and recovery in barley: key gene networks and retrotransposon response. Plant Science, 14. DOI=10.3389/fpls.2023.1193284
2022
Dueri, S., Brown, H., Asseng, S., Ewert, F., Webber, H., George, M., Craigie, R., Guarin, J.R., Pequeno, D.N.L., Stella, T., Ahmed, M., Alderman, P.D., Basso, B., Berger, A.G., Mujica, G.B., Cammarano, D., Chen, Y., Dumont, B., Rezaei, E.E., Fereres, E., Ferrise, R., Gaiser, T., Gao, Y., Garcia-Vila, M., Gayler, S., Hochman, Z., Hoogenboom, G., Kersebaum, K.C., Nendel, C., Olesen, J.E., Padovan, G., Palosuo, T., Priesack, E., Pullens, J.W.M., Rodríguez, A., Rötter, R.P., Ramos, M.R., Semenov, M.A., Senapati, N., Siebert, S., Srivastava, A.K., Stöckle, C., Supit, I., Tao, F., Thorburn, P., Wang, E., Weber, T.K.D., Xiao, L., Zhao, C., Zhao, J., Zhao, Z., Zhu, Y., and Martre, P. (2022). Simulation of winter wheat response to variable sowing dates and densities in a high-yielding environment. Journal of Experimental Botany, 73, 5715–5729.
Bretani, G., Shaaf, S., Tondelli, A., Cattivell, L., Delbono, S., Waugh, R., Thomas, W., Russell, J., Bull, H., Igartua, E., Casas, A. M., Gracia, P., Rossi, R., Schulman, A. H., and Rossini , L. (2022). Multi-environment genome-wide association mapping of culm morphology traits in barley. Frontiers in Plant Science, 13, 926277.
Sayed, M. A., Maurer, A., Schmutzer, T., Schnurbusch, T., Börner, A., Hansson, M., Pillen, K., and Youssef, H.M. (2022). Genome-wide association study of salt tolerance-related traits during germination and seedling development in an intermedium-spike barley collection. International Journal of Molecular Sciences, 23, 11060.
Senapati, N., Semenov, M., Halford, N., Hawkesford, M., Asseng, S., Cooper, M., Ewert, F., Ittersum, M., Martre, P., Olesen, J., Reynolds, M., Rötter, R. P., and Webber, H. (2022). Global wheat production could benefit from closing the genetic yield gap. Nature Food, 3, 532–541.
2021
Fernández-Calleja, M., Casas, A. M., and Igartua, E. (2021). Major flowering time genes of barley: allelic diversity, effects, and comparison with wheat. Theoretical and Applied Genetics, 134, 1867–1897.
Laux, P., P. Rötter, R., Webber, H., Dieng, D., Rahimi, J., Wei, J., Faye, B., Srivastava, A. K., Bliefernicht, J., Adeyeri, O., Arnault, J., and Kunstmann, H. (2021). To bias correct or not to bias correct? An agricultural impact modelers’ perspective on regional climate model data. Agricultural and Forest Meteorology, 304-305, 108406.
Palosuo, T., Hoffmann, M. P., Rötter, R. P., and Lehtonen, H. S. (2021). Sustainable intensification of crop production under alternative future changes in climate and technology: The case of the North Savo region. Agricultural Systems, 190 103135.
2020
Tao, F., Palosuo, T., Rotter, R. P., Hernandez Diaz-Ambrona, C. G., Minguez, M. I., Semenov, M. A., Kersebaum, K. C., Cammarano, D., Specka, X., Nendel, C., Srivastava, A. K., Ewert, F., Padovan, G., Ferrise, R., Martre, P., Rodriguez, L., Ruiz-Ramos, M., Gaiser, T., Hohn, J. G., Salo, T., Dibari, C., and Schulman, A. H. (2020). Why do crop models diverge substantially in climate impact projections? A 1. comprehensive analysis based on eight barley crop models. Agricultural and Forest Meteorology, 281, 107851.
Thorogood, R., Mustonen, V., Aleixo, A., Aphalo, P., Asiegbu, F., Cabeza, M., Cairns, J., Candolin, U., Cardoso, P., Eronen, J., Hällfors, M., Hovatta, I., Juslén, A., Kovalchuk, A., Kulmuni, J., Kuula, L., Mäkipää, R., Ovaskainen, O., Pesonen, A.-K., and Vanhatalo, J. (2020). Understanding and applying biological resilience, from genes to ecosystems. DOI:10.32942/osf.io/grpxa
Project webpage: http://www.barleyhub.org/projects/barista/
Twitter: https://twitter.com/BaristaFor
News/newsletters: