Knowledge-driven genomic predictions for sustainable disease resistance in wheat
Source: Morten Lillemo
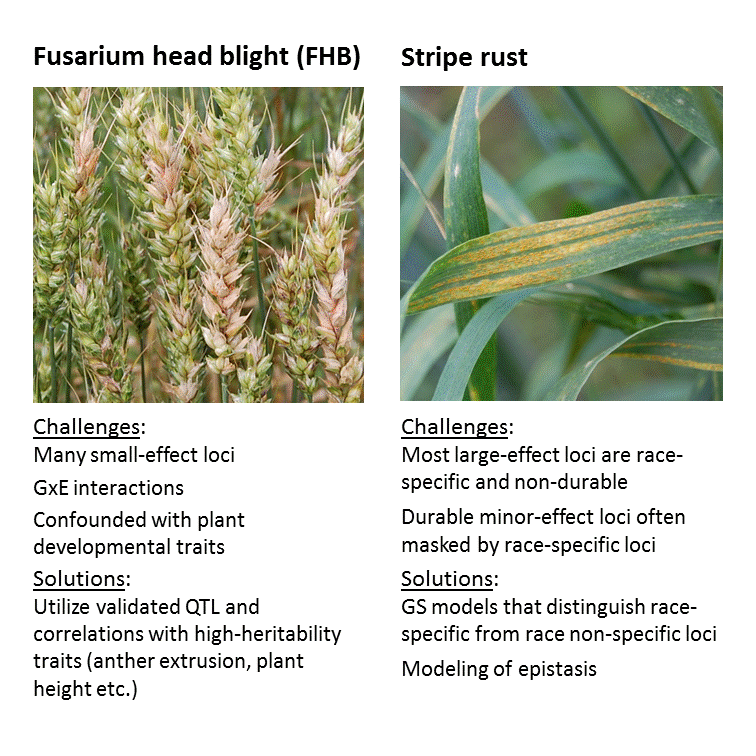
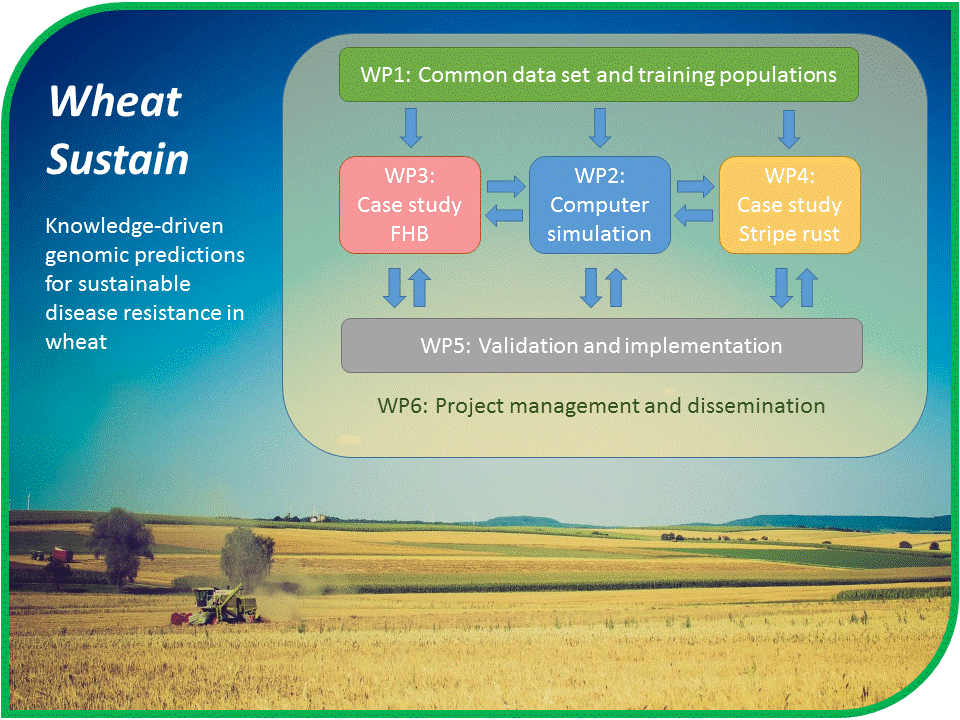
Genomic selection (GS) allows for a faster and more effective breeding of new cultivars by utilizing high-density marker data to predict breeding values of progeny lines. Depending on the prediction accuracy, time and money can be saved, and selection intensity increased by avoiding costly and time-consuming phenotyping. Much room exists to improve the genomic prediction of complex disease resistances such as stripe rust and Fusarium Head Blight (FHB) in wheat. The core idea behind WheatSustain is to make use of biologically relevant data, known trait correlations, environmental effects and quantitative trait loci (QTL) to make better and more robust GS models. In the long run, this will enable breeders to develop new cultivars with improved disease resistance, which will suffer less yield and quality losses and can be cultivated with less use of fungicides.
WheatSustain has established a close collaboration among world leading experts on genomic prediction modeling, bioinformatics, wheat genomics and leaders in the field of plant pathology and host-pathogen relationships for stripe rust and FHB resistance in wheat. The project has established an interdisciplinary teamwork across research groups from Norway, Ireland, Germany, Austria, Mexico, USA and Canada in close collaboration with public and private wheat breeding programs. Plant breeders take active part in the research by providing germplasm with phenotypic and genotypic data, conducting field trials and validating the improved GS models in their breeding programs.
Source: Morten Lillemo
As of September 2020, we have just completed the first field season with the common winter wheat training population based on germplasm from the collaborating breeders. This common training population will be used for the training of prediction models and testing new concepts of genomic prediction. The multi-location testing across sites in Germany, Austria and Norway in 2020 resulted in a high quality multi-trait data set for FHB resistance while stripe rust data with sufficient quality was obtained only from one location due to nonconductive environmental conditions for disease development at most locations. Hence, these stripe rust trials will be repeated in 2021. Aside from collecting field data on the common training population, we have used historical FHB and stripe rust data for testing our genomic prediction concepts. This has confirmed the hypothesis that multi-trait models incorporating another extrusion and temperature at anthesis can improve the prediction accuracy for FHB resistance. Likewise, historical stripe rust data has been used to test the prediction accuracies of different modelling concepts across years and inoculation methods. For validation work, new breeding lines from the collaborating breeding companies were tested for FHB and stripe rust resistance in the field in 2020 and genotyped. We have also developed statistical methods to harmonize phenotypic and genotypic data from different sources. Data harmonization enables cross-national and international comparative research, and facilitates extraction of new scientific knowledge that can lead to more optimal design of experiments. Results from the project were presented at two field days in 2020, in Austria and Germany. Other planned dissemination activities had to be cancelled due to covid-19. Campus shutdowns also caused delays in some of the lab and field activities but did not severely hamper the overall progress of the project.

Source: Morten Lillemo
Dr Morten Lillemo
Norwegian University of Life Sciences, Faculty of Biosciences, NORWAY
Email: morten.lillemo@nmbu.no
Prof Hermann Buerstmayr
University of Natural Resources and Life Sciences Vienna, Agrobiotechnology Tulln (IFA-Tulln), AUSTRIA
Dr Lorenz Hartl
Bavarian State Research Center for Agriculture, Institute for Crop Science and Plant Breeding, GERMANY
Dr Julio Isidro-Sánchez
University College Dublin, Animal and Crop Science, IRELAND
Dr Curt McCartney
Agriculture and Agri-Food Canada, Morden Research and Development Centre, CANADA
Dr Josef Holzapfel
SECOBRA Saatzucht GmbH, GERMANY
Dr Anja Hanemann
Saatzucht Josef Breun GmbH & Co. KG, GERMANY
Dr Deniz Akdemir
StatGen Consulting, NY, USA
Dr Muath Alsheikh
Graminor AS Research and Development, NORWAY
Dr Jose Crossa
International Maize and Wheat Improvement Center (CIMMYT), Biometrics and Statistics Unit, MEXICO
Mr Johann Birschitzky
Saatzucht Donau GesmbH & CoKG, AUSTRIA
2023
Garcia-Abadillo, J., Morales, L., Buerstmayr, H., Michel, S., Lillemo, M., Holzapfel, J., Hartl, L., Akdemir, D., Carvalho, H. F., and Isidro-Sánchez, J. (2023). Alternative scoring methods of fusarium head blight resistance for genomic assisted breeding. Frontiers in plant science, 13, 1057914.
Shahinnia, F., Mohler, V., and Hartl, L. (2023). Genome-Wide Association Mapping of Resistance to Warrior (-) Yellow Rust Race at the Seedling Stage in Current Central and Northern European Winter Wheat Germplasm. Plants, 12 (3), 420.
2022
Lopez-Cruz, M., Dreisigacker, S., Crespo-Herrera, L., Bentley, A. R., Singh, R., Poland, J., Shrestha, S., Huerta-Espino, J., Govindan, V., Juliana, P., Mondal, S., Pérez-Rodríguez, P., and Crossa, J. (2022). Sparse kernel models provide optimization of training set design for genomic prediction in multiyear wheat breeding data. The plant genome, 15 (4), e20254.
Montesinos López, O. A., Mosqueda González, B. A., Palafox González, A., Montesinos López, A., and Crossa, J. (2022). A General-Purpose Machine Learning R Library for Sparse Kernels Methods With an Application for Genome-Based Prediction. Frontiers in genetics, 13, 887643.
Montesinos-López, O. A., Gonzalez, H N., Montesinos- López, A., Daza-Torres, M., Lillemo, M., Montesinos-López, J. C., and Crossa, J. (2022). Comparing gradient boosting machine and Bayesian threshold BLUP for genome-based prediction of categorical traits in wheat breeding. The Plant Genome, 15 (3), e20214.
Montesinos-López, O. A., Montesinos-López, A., Cano-Paez, B., Hernández-Suárez, C. M., Santana-Mancilla, P. C., and Crossa, J. (2022). A Comparison of Three Machine Learning Methods for Multivariate Genomic Prediction Using the Sparse Kernels Method (SKM) Library. Genes, 13 (8), 1494.
Nannuru, V. K. R., Windju, S. S., Belova, T., Dieseth, J. A., Alsheikh, M., Dong, Y., McCartney, C. A., Henriques, M. A., Buerstmayr, H., Michel, S., Meuwissen, T. H. E., and Lillemo, M. (2022). Genetic architecture of fusarium head blight disease resistance and associated traits in Nordic spring wheat. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik, 135 (7), 2247–2263.
Shahinnia, F., Geyer, M., Schürmann, F., Rudolphi, S., Holzapfel, J., Kempf, H., Stadlmeier, M., Löschenberger, F., Morales, L., Buerstmayr, H., Sánchez, J. I. Y., Akdemir, D., Mohler, V., Lillemo, M., and Hartl, L. (2022). Genome-wide association study and genomic prediction of resistance to stripe rust in current Central and Northern European winter wheat germplasm. Theoretically Applied Genetics, 135, 3583–3595.
2021
Akdemir, D., Rio, S., and Isidro y Sánchez, J. (2021). TrainSel: An R Package for Selection of Training Populations. Frontiers in Genetics, 12, 655287.
Isidro Y Sánchez, J., and Akdemir, D. (2021). Training Set Optimization for Sparse Phenotyping in Genomic Selection: A Conceptual Overview. Frontiers in plant science, 12, 715910.
Montesinos-López, O. A., Montesinos-López, A., Mosqueda-González, B. A., Bentley, A. R., Lillemo, M., Varshney, R. K., and Crossa, J. (2021). A New Deep Learning Calibration Method Enhances Genome-Based Prediction of Continuous Crop Traits. Frontiers in genetics, 12, 798840.
Morales, L., Michel, S., Ametz, C., Dallinger, H. G., Löschenberger, F., Neumayer, A., Zimmerl, S., and Buerstmayr, H. (2021). Genomic signatures of selection for resistance to stripe rust in Austrian winter wheat. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik, 134 (9), 3111–3121.
2020
Akdemir, D., Knox, R., and Isidro Y Sánchez, J. (2020). Combining Partially Overlapping Multi-Omics Data in Databases Using Relationship Matrices. Frontiers in plant science, 11, 947.